1. Pre-mRNA Splicing and Fidelity Control
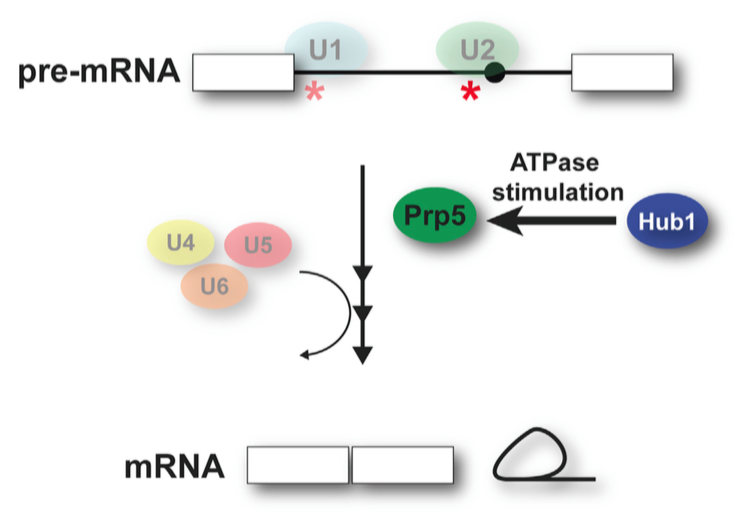
Accurate pre-mRNA splicing is needed for correct gene expression and relies on faithful splice site recognition. In our laboratory, we are interested in how splicing fidelity is regulated. We have shown that the ubiquitin-like protein Hub1 binds to the DEAD-box helicase Prp5, a key regulator of early spliceosome assembly, and stimulates its ATPase activity thereby enhancing splicing and relaxing fidelity. High Hub1 levels largely enhance splicing efficiency but also come at a cost of missplicing by tolerating suboptimal splice sites and branchpoint sequences.
In our lab, we use the budding yeast Saccharomyces cerevisiae as our main model to study. We utilize biochemical approach coupled with molecular biology and yeast genetics to understand the underlying mechanism of how splicing fidelity is regulated.
2. Potential Treatment for Clostridium difficile
In collaboration with Dr. Surang Chankhamhaengdecha from the Department of Biology, we are developing novel techniques to treat the multidrug-resistant human pathogen Clostridium difficile by using recombinant protein expression and protein engineering approaches.
