| DNA, the repository of genetic information, is under a constant attack by damaging agents from cellular metabolism and from the environment. If left uncorrected, DNA damage can lead to mutations that are deleterious to the cell. To counteract these DNA lesions, the cells have evolved several DNA repair pathways to recognize and remove the lesion, and subsequently replace it with undamaged DNA. Among the DNA repair pathways available to the cell, nucleotide excision repair (NER) is distinguished by its ability to recognize and repair a wide variety of chemically and structurally unrelated DNA lesions. In bacteria, the initial steps of NER are carried out by three proteins: UvrA, UvrB, and UvrC (Figure 1). The UvrA•UvrB complex recognizes damaged DNA and recruits UvrC, the endonuclease, to make incisions around the DNA damage. Although the Uvr proteins were among the first DNA repair proteins to be discovered, how they function together to recognize and repair a vast array of different DNA damage remains to be elucidated. |
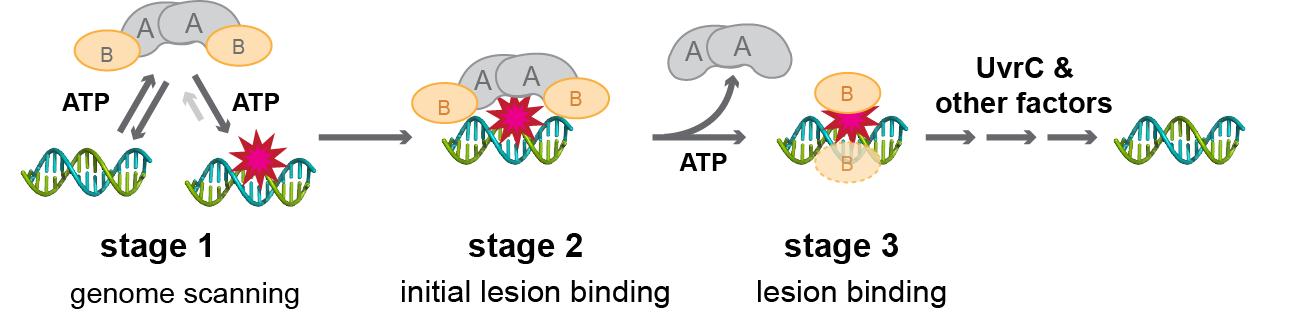 Figure 1 Overall mechanism of NER. |
| In our recent structural studies of UvrA and UvrA•UvrB complex, we showed that the UvrA protein can adopt ‘open’ and ‘closed’ conformations that we propose could underlie the mechanism of discrimination between damaged and undamaged DNA (Figure 2). Next, by using a combination of biochemical, biophysical, and structural approaches, we will investigate how interconversion of UvrA between the ‘open’ and ‘closed’ conformations could play a role in DNA damage recognition, and how such conformational changes might be regulated by nucleotide and by UvrB. DNA repair is a fundamental process that is found in all organisms. Understanding of NER is important as it plays a crucial role in the maintenance of genome integrity required for cellular processes such as transcription and DNA replication. |
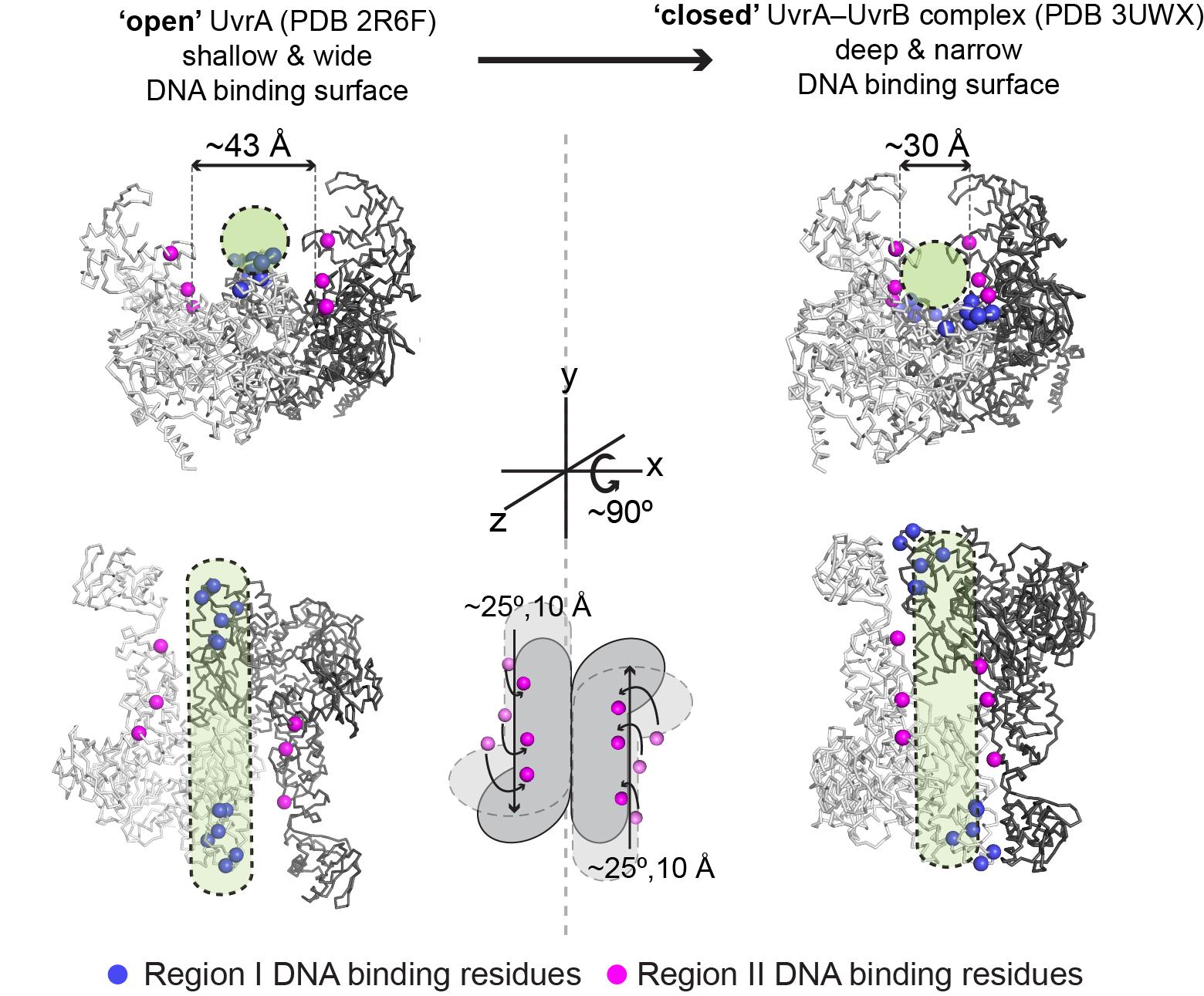 Figure 2 Model for discrimination between damaged and undamaged DNA by UvrA. Upon conformational change from ‘open’ (left) to ‘closed’ (right), the DNA binding groove is narrowed and additional residues (pink) come into contact with DNA (green cylinder). |
